
Gene expression in eukaryotes (cells with DNA inside a nucleus) and prokaryotes (single-celled organisms without a nucleus) describes how certain proteins are manufactured in specific cells according to a DNA-based recipe. Certain cell components can read gene sequences to synthesize amino acid chains (polypeptides) and proteins. Gene expression not only looks at the amount and type of proteins produced but also at when and how they occur.

Gene expression is essential to all forms of life. The genes in our DNA control every aspect of our bodies, from how we look to how we function. Without gene expression, no organism can live.
Gene expression is a synonym for messenger RNA expression or protein expression. You can learn more about protein synthesis on the associated page on this site.
A shorter explanation revolves around two processes that occur within a cell:
Copies are made of gene sequences on a strand of DNA by messenger RNA (mRNA); in eukaryotes, these ‘transcripts’ then leave the cell nucleus.
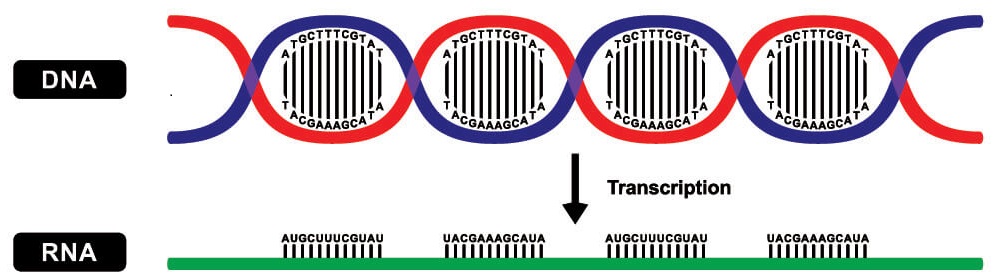
Ribosomes translate these transcripts to produce protein chains from single amino acids; they connect amino acids in the correct order according to the instructions of the copied gene sequence.

When a gene is used within a cell to produce a specific protein, that cell is carrying out gene expression. Not all cells express the same genes. Liver cells (hepatocytes) express different genes than the cells of the iris of the eye, for example.
Different tissues and organs express different genes that give them specific forms and functions. However, a complete copy of DNA is present inside every cell nucleus. Genetic engineers are quickly learning how to turn gene expression on and off in different cell types.
Genes do not code for fats or carbohydrates – only for polypeptides and proteins. Every cell relies on proteins and shorter polypeptide chains to function, regenerate, and reproduce. Even so, only about 1% of DNA contains codes for proteins. The rest is non-coding DNA that regulates how, when, and where these proteins are produced. Non-coding DNA is also known to produce RNA and the structures at the ends of chromosomes (telomeres) that protect central portions of DNA from damage.

So if you are asked, “What is gene expression?”, you need to talk about protein and polypeptide synthesis according to a specific gene found in DNA that has been turned on for that particular cell. If a gene is not expressed, it is still present within that cell’s DNA as every cells contains the complete recipe book. It is also possible for certain genes to be expressed temporarily, such as during periods of growth and development.
Gene expression steps, as already mentioned, can be found in more detail on the protein synthesis page. Any gene that codes for an amino acid sequence that produces a polypeptide chain or a protein is called a structural gene.
The below image shows a codon wheel. Messenger RNA codons are comprised of a combination of three of the following nucleobases:
A broad set of combinations give codes for one or more amino acids, as well as for start and stop signals implemented during the translation process. For example, AAA – starting from the center of the codon wheel and moving outward – codes for the amino acid lysine (Lys).

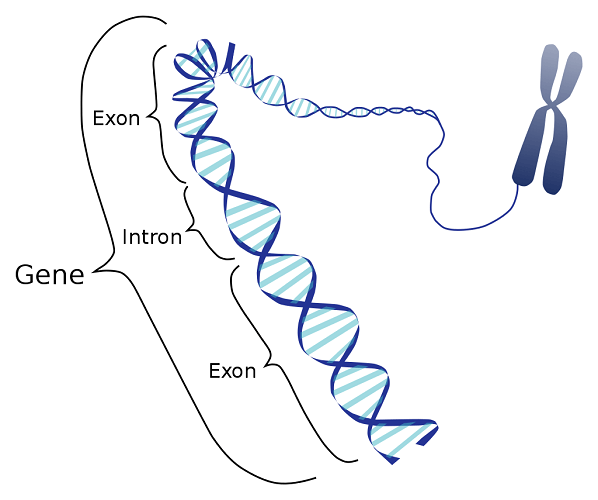
Structural genes have various components:
Gene expression is specific to the transcription and translation of DNA gene sequences in eukaryotes and prokaryotes. While eukaryotic gene expression happens inside and outside of the cell nucleus in two distinct stages, prokaryotic gene expression occurs nearly simultaneously in free-floating DNA within the cell cytoplasm.
The following four transcription steps describe eukaryotic processes.
Translation is the follow-on step from transcription; it is also composed of four similarly-named gene expression steps:
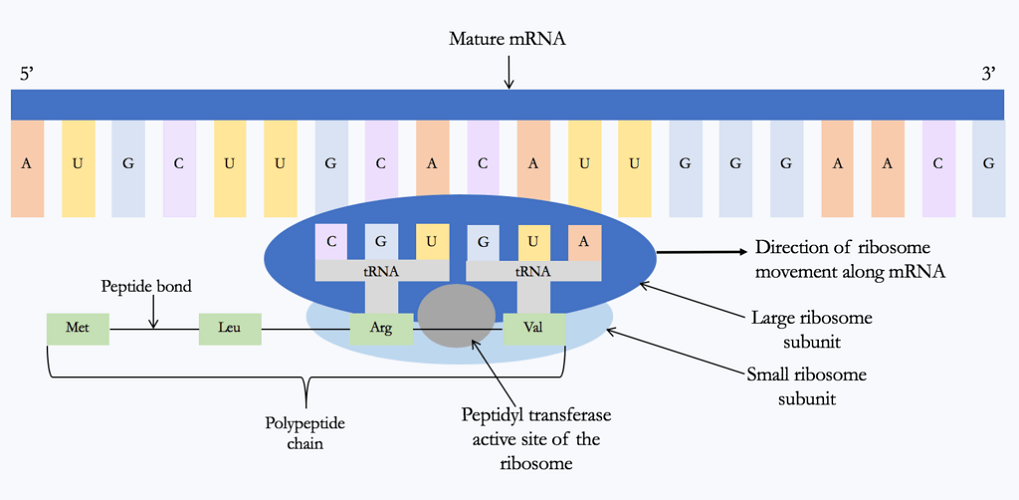
Eukaryotic versus prokaryotic gene expression uses similar steps but is not identical. This has much to do with differences in the complexity of these two cell types.
Prokaryotic organisms are single-celled and do not contain a nucleus. Instead, DNA is found within the cytoplasm. While eukaryotic cells have distinct transcription and translation phases inside and outside of the nucleus respectively, gene expression in prokaryotes happens almost in a single phase.
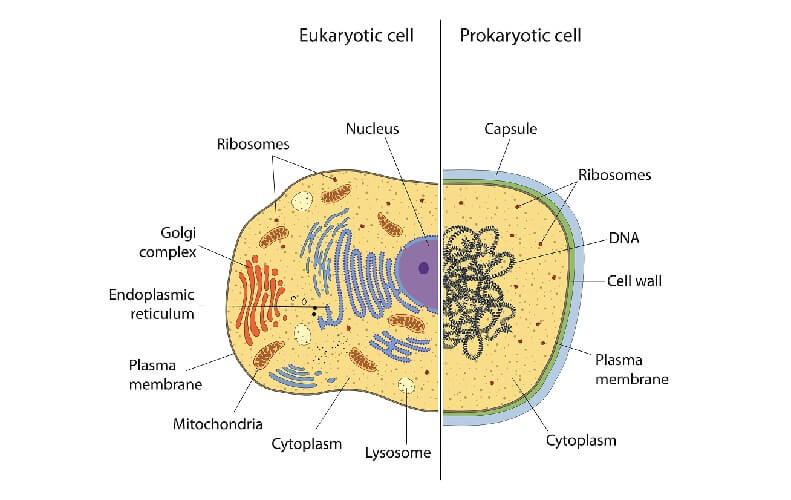
Prokaryotes only use one type of RNA polymerase that contains different molecules called sigma factors. Specific sigma factors recognize and bind to the promoters of specific genes. This is an almost self-regulating process that occurs during the transcription phase. In comparison, eukaryotes implement three different types of RNA polymerases: I, II, and III. These are regulated during both the transcription and translation phases by non-coding DNA and enzymatic transcription and translation factors.
Bacteria have the capacity to produce more than one protein from a single strand of mRNA: prokaryote mRNA can be polycistronic. Eukaryotic cells only produce one protein from one mRNA molecule – eukaryote mRNA is always monocistronic.
Prokaryotic DNA is much less complex and does not include as many repetitions as eukaryotic DNA. It is rare for bacterial DNA to contain introns. Prokaryotes also have less non-coding DNA: 95% as opposed to the 99% of eukaryotic cells. This is probably due to the self-regulatory aspect of prokaryotic RNA polymerase.
Gene expression analysis describes how scientists study gene transcription and translation to form RNA and functional proteins. Subtopics of gene expression analysis include gene cloning, epigenetics, and mutational analysis.
Looking at how certain proteins are produced by specific cell types can be done at the transcriptional, translational, and post-translational processing phases to help us to understand growth, health, and disease. Gene expression analysis uses a broad range of laboratory-based techniques to look at four main areas:
By copying transcribed RNA thousands of times (gene amplification), genetic engineers can look at how a protein or polypeptide occurs, is formed, and functions inside and outside of the cell. This type of research is most often used to compare gene expression between different samples (differential gene expression). A cancer cell that multiplies continuously might be the result of dysfunction in tumor suppressor genes, for example. By finding out exactly where this process goes wrong it may one day be possible to prevent this cause of tumor growth.

All researchers consult a gene expression database when looking into a particular gene. There are many bioinformatics sources available such as BarleyBase (plant genomics), EMAGE (Edinburgh Mouse Atlas of Gene Expression), SAGEmap (gene expression of specific nucleotide sequences), and dbERGE II (gene expression database of Experimental Results on Gene Expression). One of the largest public gene expression repositories is GEO (the Gene Expression Omnibus).
The vastness of this topic has lead to open sharing between researchers. Because of this, the field of genetic research is moving forward at a rapid pace. Although we still know very little about the complexities of gene expression, the Human Genome Project that maps the genetic blueprint of a human being was completed as far back as 2003. We now know what genes are involved in creating a human being. However, we are only just beginning to discover the genetic processes involved in human development and function.
